This research was conducted under the mentorship of Professor Dr. Dimitris Papamichail at The College of New Jersey.
This project was supported in part by the Computing Research Association for Widening Participation (CRA-WP), funded by NSF, granted to Shm Almeda and Madeline Febinger for the academic year 2018-19 as part of their Collaborative Research for Undergraduates Experience (CREU Program), as well as a Mentored Undergraduate Summer Experience (MUSE) grant from the School of Science at The College of New Jersey.
When ordering oligonucleotides for a protein variant library, there are many factors to take into account. We developed an algorithm to determine the breakpoints and overlaps for oligonucelotides that minimizes total cost. We also developed algorithms that generate useful data sets.
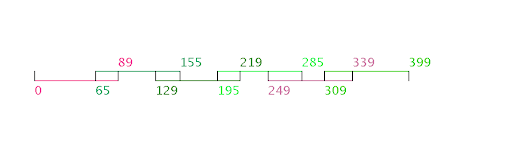
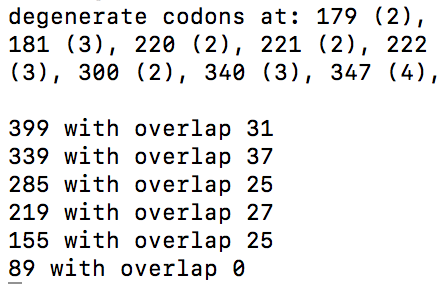
We attempt to make this work accessible and useful to the greater scientific community by developing Oligo Friend, a web tool that implements our algorithm and an online database with results generated by our programs.
Features:
Supports file upload or text input of a peptide sequence in FASTA format
Calculates the optimal breakpoints and overlaps
of oligos such that the total cost is minimal
of oligos such that the total cost is minimal
Visual and textual representation of algorithm results